REDToL: Phylogenetic and Genomic Approaches to Reconstructing the Red Algal (Rhodophyta) Tree of Life.
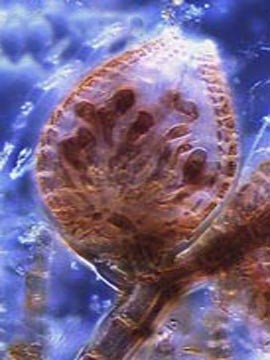
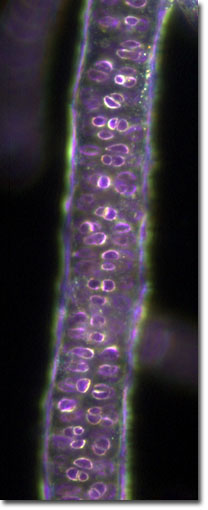
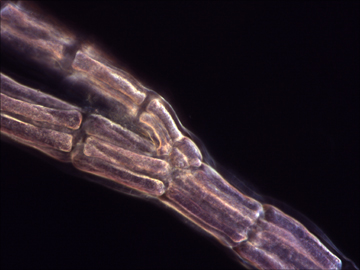
Rhodophyta (red algae) are key members of aquatic environments and sources for important human foods and have a multitude of pharmaceutical and industrial uses. Rhodophyta played an important role in the evolution of our planet through secondary endosymbiosis. A red alga was the ancient donor of the plastid in groups such as dinoflagellates, brown algae, and diatoms (among others).
Our project is aiming to: 1) reconstruct a robust phylogeny of 471 red algal species using a concatenated dataset of 2 nuclear, 4 plastid, and 2 mitochondrial encoded gene markers, 2) sequence plastid genomes and generate transcriptome databases for 16 key taxa that represent the phylogenetic (e.g., class- and order-level) breadth of the red algae, 3) make freely available red algal multi-gene and genome data via release to GenBank and a project-specific web site. The robust phylogenetic framework resulting from our study will be the basis for a comprehensive taxonomic revision of the red algae and provide the basis for interpreting key innovations during red algal evolution.
Click Tree of Life of Red Algae for more information
These Rhodophyta and Chlorophyta AToL projects will integrate data with other AToL projects (heterokonts, liverworts and other embryophytes) to identify core genes for broad phylogenetic analyses. These investigations will also provide a common framework for a future comprehensive eukaryotic tree.